About Genome Properties¶
Genome properties (GP) is an annotation system whereby functional attributes can be assigned to a genome, based on the presence of a defined set of protein family markers within that genome. For example, a species can be proposed to synthesise proline if it can be shown that the genome for that species encodes all the necessary proteins required to carry out the various biochemical steps in the proline biosynthesis pathway.
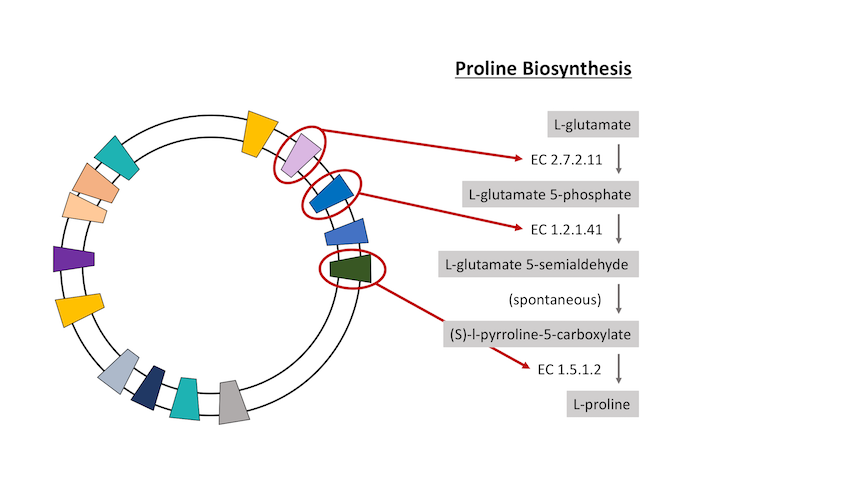
While it is possible to infer this kind of information through analysis of genomic sequence, using protein family models, like those utilised by InterPro, reduces the number of calculations while at the same time increasing the sensitivity of the query. Integrating the GP annotations into InterPro makes the process of calculating a genome property for any given genome/proteome faster and more accurate than by sequence comparison. The process is further streamlined by the fact that all UniProt sequences, already come with InterPro matches calculated. Each GP is defined as a numbered series of biochemical steps, which in turn have some form of evidence determining the presence of the protein required for that step (usually a profile hidden Markov model, or HMM). The property may include steps which are not strictly necessary, but often take place within the process being described. In the calculation of a GP, the evidence for each required step is tested against the genome/proteome under query. Where all steps can be encoded by the query genome, the output is a YES. Where some (above a defined threshold level) steps can be shown to be encoded, the output is a PARTIAL. Where no steps can be shown to be encoded (or fewer than the threshold level), the output is a NO.
For more information about calculating Genome Properties, see here.
How to access Genome Properties data¶
Browse by Genome Property: You can browse to your GP of interest using the hierarchy browser. GPs are arranged into categories and subcategories for easy naviagtion. Each GP contains a descriptive abstract describing the property, as well as the individual steps comprising it. For more infomration on the content of the GP files, see here (link to flatfile description).
You can also navigate to your GP of interest using the lists of various property types (PATHWAY, SYTEM, GUILD, etc) provided under the Browse tab.
Browse by Proteome/Genome: You can browse GPs by species/genome/proteome using our customisable viewer. All GPs are calculated against a representative set of proteomes as provided by UniProt. The output of each GP calculation (yes, no or partial) is reported for each species, in the form of a colour coded matrix. This viewer is easily customisable by the user to allow only specified species/proteoms of interest, as well as GPs of interest, to be included and compared. This viewer allows you to quickly and easily compare the overall “fingerprint” of GP content for a set of species, or indeed the species distribution of a set of GPs.
Upload your data: Further to the features of our interactive viewer already described, you are also able to upload your own proteome data and compare this against the representative set of proteomes available. By analysing your proteome of interest using InterProScan, an output file of InterPro matches is produced. This file (tsv version) can be uploaded to the viewer page, allowing you to view the GP results for your proteome of interest in the colour-coded matrix viewer alongside your chosen comparison set of proteomes/species.
Background¶
Genome Properties were developed at J. Craig Venter Institute within the TIGRFAM group [PMID:15347579, PMID:23197656] as a way to improve the functional annotation of proteins, as well as providing a resource to assist in comparative genomics. They were based predominantly on TIGRFAM HMM models for step evidence, supplemented with some Pfam models.
One of the main benefits of integrating GP into InterPro is that through the InterPro system of pulling together protein signatures from a range of member databases, there is now a very large potential pool of protein models available to use as evidence for steps, including multiple levels of specificity. In general, we use specific family models in the calculation of GP steps.